Overview
This vignette demonstrates how to create a time series of NDVI (Normalized Difference Vegetation Index) from NASA’s Harmonized Landsat Sentinel-2 (HLS) data. NDVI is a widely-used vegetation index that measures the “greenness” of vegetation.
We’ll cover:
- Querying HLS satellite data from STAC
- Creating virtual rasters and applying cloud masks
- Computing NDVI from spectral bands
- Applying temporal filtering to reduce noise
- Creating an animated GIF visualization
Setup
First, load the package and set up parallel processing. We’ll use 6 workers to speed up data processing:
Step 1: Define Your Area of Interest
We’ll create a bounding box around a point in West Texas, extending it to cover a reasonable study area:
# Create a bounding box around a point in West Texas
# The point is at -102.2°W, 33.1°N
# extend_x and extend_y control the size of the area (in degrees)
bbox <- gdalraster::bbox_from_wkt(
wkt = "POINT (-102.2 33.1)",
extend_x = 0.25,
extend_y = 0.125
)
# Project the bounding box to Azimuthal Equidistant projection
# This projection preserves distances from the center point
te <- bbox_to_projected(bbox, proj_generic = "aeqd")
trs <- attr(te, "wkt") # Extract the projection WKT stringWhy project? Projecting to Azimuthal Equidistant ensures accurate distance measurements from the center point and proper area calculations for your study region.
Step 2: Query HLS Data from STAC
Query NASA’s HLS STAC catalog for Sentinel-2 data over your area and time period and request only the bands we need for NDVI calculation (B04;Red and B08; Broad Near Infrared) and cloud masking (Fmask):
# Query HLS Sentinel-2 data for 2021
tex_s2 <- hls_stac_query(
bbox,
start_date = "2023-12-15",
end_date = "2024-12-30",
# Request specific spectral bands and the quality mask
assets = c("B04", "B08", "Fmask"),
max_cloud_cover = 30, # Allow up to 40% cloud cover
collection = "hls2-s30" # HLS Sentinel-2 at 30m resolution
)
# Check how many images were found
cli::cli_alert_info(sprintf("Found %d HLS scenes", rstac::items_length(tex_s2)))
#> ℹ Found 170 HLS scenesStep 3: Create Virtual Rasters with Cloud Masking
Build a virtual raster collection from the STAC results, applying cloud masking and warping to your target projection/resolution:
tex_s2_collect <- vrt_collect(tex_s2) |>
# Apply cloud/shadow/water mask using Fmask band
# Values 0-3 represent clear conditions
vrt_set_maskfun(
mask_band = "Fmask",
mask_values = c(0, 1, 2, 3), # 0=Cirrus, 1=Cloud, 2=Adjacent to cloud/shadow, 3=Cloud shadow
build_mask_pixfun = build_bitmask()
) |>
# Warp all images to common projection and resolution
vrt_warp(
t_srs = trs, # Target projection
te = te, # Target extent
tr = c(30, 30) , # Target resolution (30m pixels)
lazy = FALSE
)Key concepts:
- Virtual rasters: No data is copied or resampled yet - we’re building instructions for GDAL
- Cloud masking: Pixels flagged as clouds, shadows, or other problematic features are masked out
- Warping: Ensures all images are aligned to the same grid for time series analysis
Step 4: Calculate NDVI and Apply Temporal Filtering
Set the NDVI calculation using the vrt_derived_block()
function which automatically restructures the data and sets the forumla
as a VRT pixel function using muparser expressions. This is then
computed during the call to singleband_m2m(), where the NDVI values are
temporally filtered using a Hampel filter to reduce noise from residual
clouds and sensor artifacts:
tex_ndvi <- tex_s2_collect |>
# Calculate NDVI: (NIR - Red) / (NIR + Red)
vrt_derived_block(
ndvi ~ (B08 - B04) / (B08 + B04)
) |>
# Apply Hampel filter to detect and replace outliers
# This helps remove residual cloud contamination and sensor noise
singleband_m2m(
m2m_fun = hampel_filter(
k = 3L, # Window size: 3 images before and after
t0 = 0, # Threshold: 0 standard deviations (aggressive filtering)
impute_na = TRUE # Replace no data with previous valid value
),
recollect = TRUE # Rebuild VRT after filtering
)About Hampel filtering:
The Hampel filter identifies outliers in each pixel’s time series by computing the median absolute deviation (MAD) within a sliding window of neighboring time steps. For each pixel at time t, it examines the window [t-k … t+k] and flags values that deviate more than t0 x MAD from the local median.
-
Window size (
k=3): Each pixel is compared against 7 time steps (3 before, current, 3 after) -
Threshold (
t0=0): Any value differing from the local median is flagged as an outlier (equivalent to applying a pure median filter). Settingt0=3would allow values within 3 MAD units of the median (Pearson’s rule of normal data), making the filter more forgiving. -
Imputation (
impute_na=TRUE): Outliers are replaced with the last valid (non-outlier, non-NA) value from earlier in the time series, effectively carrying forward the most recent “good” observation
This is particularly effective for removing clouds and atmospheric artifacts that Fmask missed, as these appear as sharp deviations from the local temporal pattern.
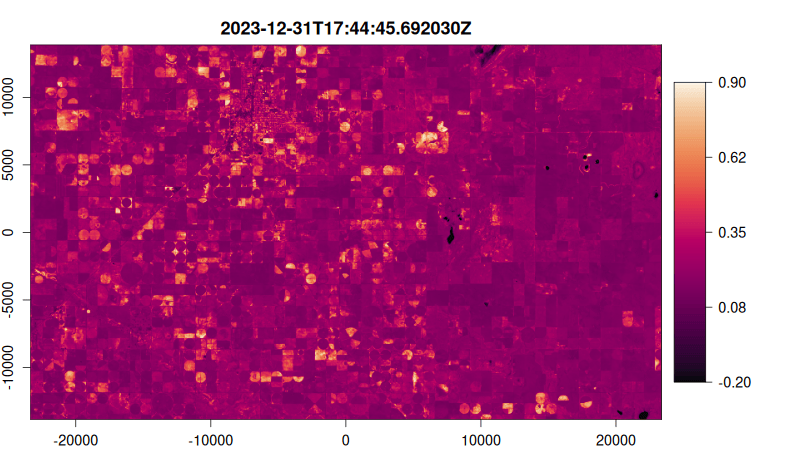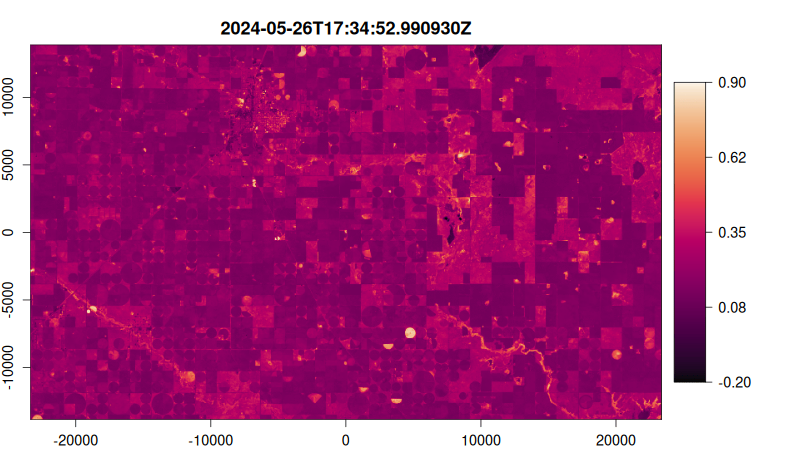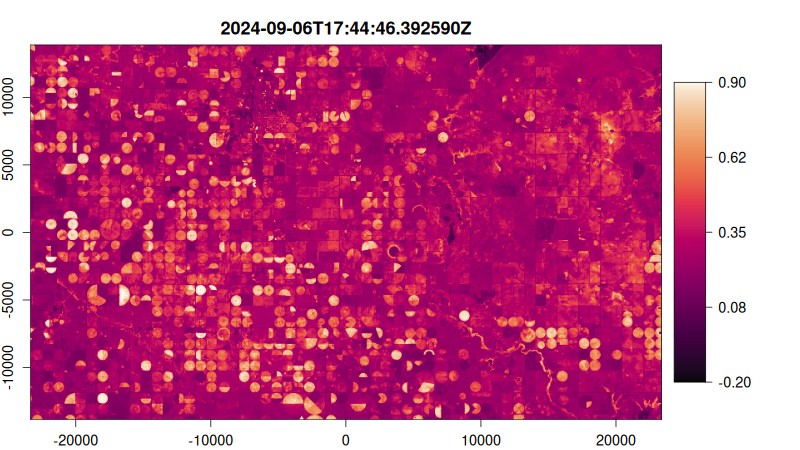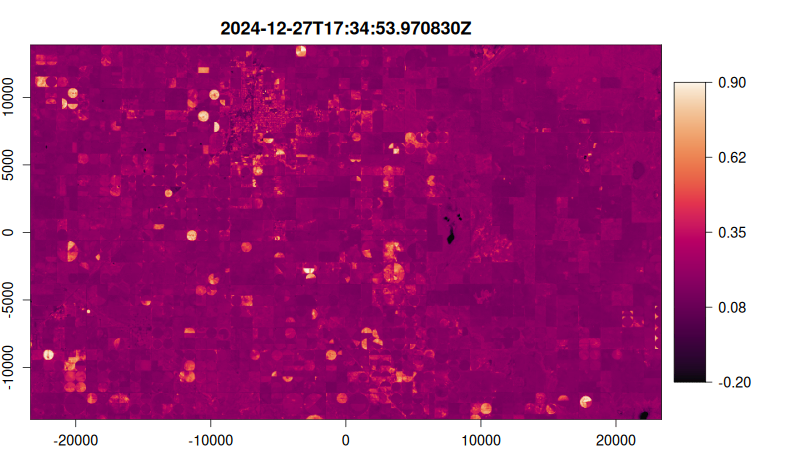
Step 5: Visualize as an Animated GIF
Create a function to plot each NDVI image, then render as an animated GIF:
# Create plotting function
ndvi_plot <- function() {
# Start from image 10 stabilise median filter effect
purrr::walk(10:tex_ndvi$n_items, function(i) {
plot(
tex_ndvi,
item = i,
col = hcl.colors(100, "Rocket"),
title = "dttm", # Use datetime as title
minmax_def = c(-0.2, 0.9), # Set NDVI color scale range
)
})
}
fs::dir_create("figure") # Create output directory if it doesn't exist
# Generate the animated GIF
gif_file <- gifski::save_gif(
ndvi_plot(),
gif_file = fs::path("figure", "ndvi_timeseries.gif"),
width = 800, # Image width in pixels
height = 450, # Image height in pixels
delay = 0.1 # Delay between frames (seconds)
)